White Paper
Interoperability between Stemmatological Microservices
Tara Andrews1, Simo Linkola2, Teemu Roos2, Joris van Zundert3
- KU Leuven (BE)
- Helsinki Institute for Information Technology HIIT, University of Helsinki (FI)
- Huygens Institute for the History of the Netherlands, Royal Netherlands Academy of Arts and Sciences (NL)
7 May 2013
In 2012 two online tools for text stemmatology research were published to the web: Stemweb [source code available at https://github.com/Stemweb/Stemweb], under development by members of the Helsinki Institute for Information Technology HIIT, and Stemmaweb [http://byzantini.st/stemmaweb/; source code available at https://github.com/tla/stemmaweb/] developed by the Tree of Texts project at KU Leuven in collaboration with members of the Interedition project [http://www.interedition.eu]. Stemweb is a resource for many sorts of phylogenetic and other statistical analysis of a text, including the RHM and SemStem methods developed specifically for the case of recovering manuscript text stemmata. Stemmaweb is a complementary resource for the visualization, regularization, and analysis of the variation within a text using graph search methods, and allows the scholar to define as many hypothetical stemmata as she or he would like to explore. On 25-26 April 2013 a meeting was held, with funding generously provided by a Small Project Grant of the European Association for Digital Humanities (ALLC), to create an open blueprint for integration. Our blueprint is meant not only to be open to the two existing tools but also to provide a framework for interoperability with any other tool for stemmatological research that may appear in future. The blueprint is provided here in the form of a white paper; comments are invited from interested developers and scholars, and those received by 31 May will be taken under consideration in time for the first implementation.
Background
Stemmatology is the study of how to derive computationally the copying relationships between ancient and medieval manuscripts. Various statistical and algorithmic approaches have been adapted from the field of evolutionary biology for this over the past few decades, such as maximum parsimony and neighbour joining; others, such as the RHM method and the more recent Semstem, have been developed specifically with the problem of text genealogy in mind. Many of the methods require the scholar to become familiar with software packages for evolutionary biology, and are in that sense not particularly approachable (or even, in the case of non-free software, not easily available.) One of the greatest advantages of Stemweb is precisely the collection of several applicable algorithms in one place, so that the scholar can use different methods on the same dataset without having to devise a different technical working process for each.
To allow practical application of and reflection on the various algorithms by as wide a community of scholars as possible, our aim is to provide both open GUI and API access to these tools. The web-based user interfaces allow the integrated facilitation of the various methods for stemmatology developed by different researchers in various locations. Founding this integration on the basis of (web) APIs will allow anyone who develops additional approaches to stemmatology to let their solutions be interoperable with the current published ones.
Where Stemweb provides a collection of algorithms for the creation of stemmatic hypotheses, Stemmaweb is a collection of tools designed to examine and analyse collated sets of texts, the relationships between variant readings, and the logical consequences of one or more stemmatic hypotheses. In this context Stemmaweb is a consumer of the hypotheses that Stemweb can generate.
Aims
The aim of this proposal is thus to allow any scholar to:
- have open web access to our current technology for stemmatology
- provide an open interoperable way to contribute stemmatological algorithms to the framework
- provide an open API that allows integration of stemmatological services in any web-enabled GUI
Throughout future stages of the project, we wish to continue an active engagement with the scholarly community concerning the direction and functionality of the ecosystem of tools for stemmatological and text-genetic research. As far as possible given the resources available to us, we will provide an open communication space for scholars to reflect, comment on, and participate in our work. We also intend to provide or collect information on the properties of different stemmatological methods as well as online guidelines for their use and other documentation.
Request for Comments
At this point in time we have provisionally agreed on the primary APIs to connect and interoperate the Stemweb and StemmaWeb solutions. We present in this white paper the proposed protocol and data formats that will allow the interconnection of these two solutions. We have striven to keep this protocol for interoperability lightweight, extensible and open so that any developer, researcher, or contributor may scrutinize, comment and suggest changes and enhancements to this protocol.
Description of the proposed framework
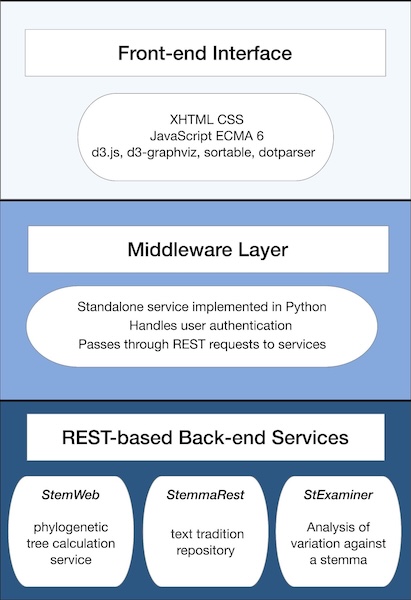
The protocol proposed is based on the idea of microservices. These are very small web services, with a RESTlike API as far as possible. The API normally uses JSON as its means of exchange and communication between server and client. Microservices have been defined in the context of the Interedition project (http://interedition.eu/wiki/index.php/About_microservices). Individual microservices may be combined in various ways to drive the functionality of multiple web applications. The interoperability of Stemweb and Stemmaweb follows this model as depicted in Figure 1.
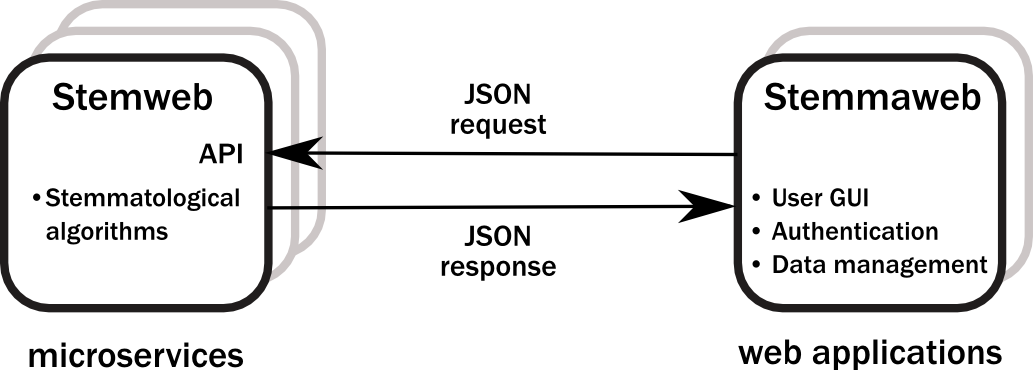
Figure 1: Microservices architecture
In this instance the Stemmaweb visualisation service will be a client of the microservice interface provided by Stemweb, sending collation data and receiving one or more stemmatic tree hypotheses in return. The aim is nevertheless to define an API whose ‘server’ functions can be implemented by any other microservice that provides a stemmatological algorithm, and the ‘client’ functions can be implemented by any consumer of such an algorithm.
Server-side API
GET /algorithms/available
which will return a JSON response that lists the stemmatology algorithms available on the server, along with descriptions for display in a user interface and option parameters that are required or recommended for their use. The response takes the following form:
[ { model: argument
pk: <ID in server database>
value: <argument value’s type, see below>
verbose_name: <human readable name of the argument>
description: <longer description of argument’s behaviour>
},
{ model: argument
...
},
...
{ model: algorithm
pk: <ID in server database>
name: <human readable name>
desc: <longer description of the algorithm>
url: <link to original article, if available>
args: [<list of argument pk’s for input arguments>]
},
{ model: algorithm
...
},
...
]
Valid argument values at present are: positive_integer, integer, boolean, input_file, float and String.
POST /algorithms/calculate
The client should send the following request data to accompany the POST request:
{ userid: <email / ID of user>
algorithm: <ID of algorithm>
parameters: { ... }
data: <string containing data in specified format> }
The server will indicate its response via appropriate HTTP status codes, e.g.:
200 OK (job was accepted and will now be processed
{ jobid: id }
400 Bad Request (e.g. request was malformed)
{ error: <error message> }
403 Forbidden (e.g. client not authorized)
{ error: <error message> }
The algorithms for stemmatology calculation can take an arbitrarily long time to run, and we therefore propose an asynchronous callback method for the return of results. For this reason the initial server response to a successful request will consist only of a job ID.
When the calculation is finished, the server must make a POST request to a location implemented on the client side with the results. The return of a 200 response implies a commitment that the job will run and the results, whether success or error, will be returned. See below, ‘Client-side API’, for more information.
GET /algorithms/jobstatus?jobid=<ID number>
{ jobid: <id number>
statuscode: < 1 = running / >1 = failed / 0 = success >
*result: <data>
*result_format: <format> }
The “result” keys should only be included, where applicable, if the job is no longer running. This is intended primarily as a fallback interface in case the server was unable to make the initial POST reporting results to the client as described above.
Client-side API
In addition to ensuring that requests to the server API are well-formed as documented above, a client to the stemmatological algorithm microservice must implement a URL to which the server can post results:
POST /stemmatology/result
(Success):
{ jobid: <ID number>
statuscode: 0
result_format: <format>
result: <data> }
(Failure):
{ jobid: <ID number>
statuscode: >1
result: <error message> }
In case no results are received in a reasonable time, or it is otherwise suspected that the results of a job failed to be returned, the client may request a job status from the server as documented above.
Crediting issues
When building web services based on this framework, the author(s) should in each case ensure that the providers of the various solutions and components on which the services are built are given due credit – for instance, that a user using Stemmaweb will be notified that the stemmatological algorithms are provided by Stemweb. A general technical solution to the problem of “giving credit where credit is due” is beyond the scope of this white paper; comments on the issue are nevertheless welcome for the purpose of creating a suitable policy in the future. At this point we can only recommend that web service authors make liberal use of logos.
Implementation plan
We are presently releasing this white paper for comments on the Digital Medievalist mailing list (http://www.digitalmedievalist.org/), Humanist (http://dhhumanist.org/), and the Textual Scholarship blog (http://www.textualscholarship.eu/). Comments are invited on the initial phase until the end of May 2013. The teams at KU Leuven, HIIT, and Huygens ING will begin implementation of the functionality required by the framework and the API in June 2013, with the intent of releasing the complete and tested web services by the end of November 2013.
We hope particularly to solicit comments concerning the API from the point of view of potential future extensions of the services outlined above, such as other tools and resources (both front-end and back-end) that can be implemented in the framework. For instance, it will probably be beneficial to integrate a collation tool such as CollateX into the system, with seamless data storage or sharing between the microservices so that the user need not repeatedly upload and download his or her data in order to perform a full analysis cycle. Another desideratum might be integration with an informational resource such as the Parvum lexicon stemmatologicum [https://wiki.uib.no/stemmatology/index.php] produced by members of the Studia Stemmatologica group, which is a collection of definitions of terms used in stemmatology. The API should be able to accommodate any such extensions as seamlessly as possible.